|
Workshop by Deborah Paul for the
Southern California Association of Marine Invertebrate Taxonomists (SCAMIT), 8 August 2011 in Costa Mesa, California. This
workshop hosted by the Southern California Coastal Water Research Project (SCCWRP).
Title: SCAMIT - Morphbank Workshop: Custom Workbook Submission Author: Deborah Paul, Florida State University Abstract: Bulk Upload to Morphbank using a customized Excel workbook. SCAMIT members attending this workshop spent the morning learning how to submit and edit records uploaded to Morphbank for the SCAMIT group. Topics included how to find objects and images once they are in Morphbank, how to make a Morphbank Collection as well as updates about new fields and features of Morphbank version 3. In the afternoon, focus shifted to how to populate an Excel workbook specifically tailored to meet the needs of the SCAMIT group for bulk upload. Efforts were made to hightlight the relationships between fields in this workbook with the fields seen earlier in the morning on the Morphbank website. At the end of the workshop, one of the participant's workbooks was uploaded to the website to show the entire process. |

download presentation: pptx Aug 2011 |
|
Presentations by Deborah Paul for the
Naturhistoriska Riksmuseet Biodiversity Informatics Course, 18 September 2009 in Stockholm, Sweden.
Title: Morphbank Current Topics: Using Images and Metadata Authors: Deborah Paul & Greg Riccardi, Florida State University Abstract: Morphbank as an Image Repository & Bioinformatics Research Tool About 25 graduate students and a few other scientists participated in a 2-week Biodiversity Informatics course held in September 2009 at the Swedish Museum of Natural History in Stockholm, Sweden. Students came from various universities in Europe and South America. This course consisted of teaching the students how to create and access a simple database (using their own data on their own computers) and then various lecturers introduced the students to different software they might find useful for their research. The workshop was hands-on. The Morphbank lecture presented here acts as an overview of what Morphbank is and later in the afternoon, the students focused on how they might get data into the system. Some students uploaded data and images to Morphbank as part of their participation requirement in the workshop. Each student was expected to complete a project using some of the software they had been introduced to during the 2 week period. |

download presentation: ppt Sep 2009 |
|
Presentations by Deborah Paul for the November 2008 SCAMIT meeting in San Diego, California.
Title: SCAMIT and Morphbank Authors: Deborah Paul, Katja Seltmann & Greg Riccardi, Florida State University Abstract: A Strategy for SCAMIT to Use Morphbank as an Image Repository The Southern California Association of Marine Invertebrate Taxonomists (SCAMIT) group was formed in 1982 with the goal of promoting the study of marine invertebrate taxonomy in southern California and developing a regionally standardized taxonomy. Furthering this goal, they are in the process of designing and building a web site. The web site will include species pages for the public as well as a secure section for SCAMIT members complete with a blog and taxonomic keys. Morphbank was chosen by SCAMIT as their image repository. SCAMIT sent members to Tallahassee in May 2007 to the Morphbank Usability Study and Workshop. Since learning to use Morphbank via the web SCAMIT members would like to streamline the process of data and image submission. In collaboration with SCAMIT, Morphbank designed a simpler Excel workbook for users. Four SCAMIT meetings are planned for November 2008. The first meeting on November 17th focuses on a possible design for their database, an explanation of how the images will be called from Morphbank to appear on the SCAMIT pages and an introduction to the newly-designed, easily-customized Excel workbook for data submission. Regarding the Excel workbook, the idea is to give any Morphbank contributor a fast way to compile data for submission (as much or as little data as the user desires). In Morphbank, it is now possible to submit an image with as little metadata as a taxonomic name (can be a temporary taxonomic identifier) and a unique external identifier for the image (a user-provided GUID, LSID, etc). These changes to the Morphbank database make it possible to use Morphbank strictly as an image repository with little or no metadata regarding the specimen, view, or locality information. After sending a completed workbook to Morphbank, the contents are put in XML format for upload to Morphbank. The next 3 SCAMIT meetings, November 18th - 20th, focus on how to use this new Excel Workbook. Each Lab (LACSD, OCSD, etc.) will be submitting data for upload to Morphbank. The GUIDs needed for each image will be assigned by a single individual in each lab, or one central person SCAMIT chooses. The powerpoint explains how to populate the workbook and where to go for help if there is a question. |
download presentation: ppt Nov 17th, 2008 download presentation: ppt Nov 18 - 20, 2008 |
|
Presentation by Greg Riccardi, Austin Mast, Daniel Miranker and Ferner Cilloniz at the 2008 tdwg meeting.
Title: OntoMorphBankSter: Image-driven Ontology and/or Ontology-driven Image Annotation Authors: Greg Riccardi, Austin Mast (Florida State University), Dan Miranker, and Ferner Cilloniz (University of Texas). Abstract: We describe a method of linking images and ontologies realized as a combination of two systems: the Morphbank image and metadata repository (http://www.morphbank.net/) and the Morphster ontology illustration and management system (http://www.morphster.org/). Our formal treatment of images allowed us to create an image-driven ontology editor to enable domain experts to build ontologies as a part of the research process. Two concepts are linked in this system: image-driven ontology, which couples images with classes and properties to provide a rich definition of the concepts described, and ontology-based image annotation, which tags images with ontology terms to identify the areas of interest within the image that are described by the terms. The two ideas combined support the notion of standard view descriptions-- combination of ontology terms that describe what can be seen in an image. A group may predefine standard views and associate appropriate views with their images. This aggregation of terms into standard views makes it easy for users to give precise descriptions of their images using terms that are understood within the group. The combined system relies on standard interaction methods. Morphbank has Web services that support query, insert and update. Morphster provides interoperability among a variety of ontology systems. Morphster interacts with Morphbank services to create a dynamic linking between concepts and images.This integration is embodied in OntoBrowser (http://www.morphster.org:8080/OntobrowserV3/Ontobrowser.html). As images supplant hands-on observation of specimens in systematic research, the use of search methods to retrieve related images is a critical function. An open research question in this environment is how can semantically rich systems (i.e. ontology) exploit those semantics when accessing and retrieving images from legacy databases. In our context, OntoBrowser is a semantically rich environment. Users may choose among ontologies of different organisms. Individual images may be found by browsing by ontology terms or through a search mechanism that leverages the ontology. Ontology-based image retrieval systems use a basic definition of similarity that involves a graph-based distance measure among the set of ontological terms in the query image and the set of ontological terms associated with each image in the database. This is the same problem as measuring the similarity of the function of two genes per their annotation in the Gene Ontology. A search will be BLAST-like in the sense that the ontology annotations characterize the similarity between images. The results will be ranked based on the information that scientists have associated with the images. Our ontology activities are informed by our collaboration with the Spider Assembling the Tree of Life (SATOL) project (http://research.amnh.org/atol/files/), Phenoscape (https://www.nescent.org/phenoscape) and the Hymenoptera Ontology (http://ontology.insectmuseum.org). The attached figure displays information from SATOL. Morphbank contains the standard view manual and the images collected by SATOL. Morphster combines the SATOL ontology and images from the view manual to create an illustrated anatomy atlas. In the figure, the left panel is a browsable tree view of the anatomy ontology that shows the term spinnerets, labeled images documenting the term appear in the center panel, and other images in Morphbank tagged |

download presentation: ppt |
|
Presentation by Boyce Tankersley and Greg Riccardi at the 2008 tdwg meeting.
Title: Data Integration Services for Biodiversity Informatics: An Example from Plant Collections and Morphbank Authors: Greg Riccardi (Florida State University) and Boyce Tankersley (Chicago Botanic Garden). Abstract: An interoperable information system has been created that combines the Morphbank image repository, PlantCollections botanical information portal, and several collections management systems. Putting these systems together poses many challenges that are common in biodiversity informatics. Plans are in place to make the system compatible with the TDWG Access Protocol for Information Retrieval (TAPIR). TAPIR compatibility will allow small repositories to be information providers and to allow consumers to feed back information to providers. TAPIR is TDWG project that specifies an XML-based protocol for accessing structured data that may be stored on distributed databases of varied structure. TAPIR combines and extends features of the BioCASe and DiGIR protocols to create a new means of communication between data providers and client applications using the Internet. PlantCollections (PC) publishes information about public gardens on the Web using the Google Base database and XML technologies. PC relies on Morphbank for storing its images and uses Morphbank services to populate images on its pages. The PC project is an important educational and scientific activity that is integrating beautiful images and garden information with taxonomic information and other scientific content. Botanical gardens keep extensive records of their specimens, with taxonomy and locality information related to current location and natural habitat. This information is however not readily available outside of the individual gardens. The gardens typically have little informatics infrastructure and no ability to provide 24/7 servers. The infrastructure is therefore often insufficient to allow the gardens to become direct TAPIR providers. The major challenges for PC were to provide infrastructure to aggregate and publish the databases and the photographic collections of the gardens, and to provide for information update from garden to portal and back. The PC project began with a survey of the database fields in the garden databases, the development of a common metadata schema, the deployment of that schema using Google Base, and the transfer of many images to the Morphbank image and metadata repository. The typical information flow to publish garden data is: The garden information including image metadata is pushed to Google Base The image files are pushed to Morphbank via ftp Specimen metadata is pushed from Google Base to Morphbank via Web services The PC Portal provides an interface to access metadata and images with active support from Morphbank The interaction between garden database, Google Base, the PC Portal and Morphbank is accomplished using XML documents and Application Programming Interfaces (APIs) that are consistent with TAPIR. The full implementation of updates is not present and is not fully described in TAPIR. Two major requirements for interoperability for PC are to extend the Darwin Core, Dublin Core, and TAPIR metadata standards to include support for images and annotations, and to create a suitable update specification for biodiversity metadata. The schema for data exchange will be created from the Dublin Core and Darwin Core metadata schemas and the TAPIR schema. Minor modifications to the existing Morphbank API and schema will be necessary for conformance. The existing Morphbank services were designed to be adaptable to new schemas. The creation of these standards will be in collaboration with TDWG, GBIF and the Atlas of Living Australia. |

download presentation: ppt |
|
Presentation by Fredrik Ronquist and Greg Riccardi at the 2007 tdwg meeting.
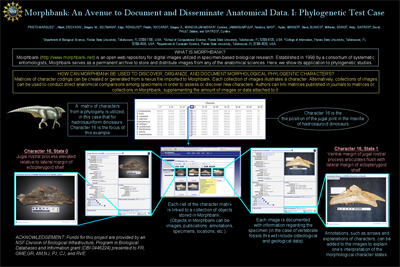
Title: Building Relationships between Images and other Biological Objects using the Morphbank Repository (www.morphbank.net) and Morphbank Collaboratory Tools Authors: Greg Riccardi, Fredrik Ronquist, Austin Mast, Neelima Jammingumpula, Katja Seltmann, Karolina Maneva-Jakimoska, Steve Winner and Deborah Paul (Florida State University), and Andrew Deans (North Carolina State University). Abstract: An overview presentation outlining the Morphbank project, its long-term goals and the present software. Includes linking and embeding images, the ITIS collaboration and incorperating ontologies into the system beginning with the Hymenoptera Ontology. Morphbank was designed to be a secure repository for images and metadata. It includes a rich data model that conforms to open community-supported standards, such as Darwin Core for specimen metadata. The metadata is available in Web pages and in XML and RDF. Access to content is managed by content owners and can be restricted to a specific group of users or made publicly available. The security model ensures that researchers can take advantage of the Morphbank system during the entire research lifecycle: from initial data collection and imaging through publication. The Morphbank team has built enabling technology for biodiversity researchers, particularly users of biodiversity research collections and morphological phylogeneticists. In the process, we have built connections to other providers of functionality (e.g., HERBIS, Specify) and biodiversity content (e.g., ITIS, the Integrated Taxonomic Information System). A major advance in the usability of Morphbank comes from collections management, which allows a user to accumulate digital objects (private and public) in collections. Users can accumulate all of the objects used in making scientific interpretations, including images, image annotations, publications, operational taxonomic units (OTU), character matrices, and even other digital object collections. The digital objects that can be included in a collection are not restricted to objects within the Morphbank system, but can be any digital object with a globally unique identifier (GUID). Collections can be published, and can form the kernel around which new collections are built. Collections provide a powerful way of documenting and publishing scientific observations and opinions. The Morphbank Collections Interface shows a collection that has been created to code a character in a matrix for phylogenetic study or the production of a multi-entry identification key. The user has organized the image tiles in order to define a particular feature. In this case, the character represents the occurrence of spots on butterfly wings. The states are many spots, few spots and no spots. Each blue tile and the thumbnails that follow it represent one of the states and some images that are coded with that state. The character can be used in one or more matrices and additional images can be coded and added to the state. This strategy for representing character matrices is designed to enhance the interaction with systems that focus on matrices, like Morphobank and Mesquite. By using the images as the primary source of character information, we provide a different look and feel to the character coding process. Morphbank is a rich resource for other systems to use in the discovery and illustration of characters and states. The presentation will emphasize the connections that have been defined by researchers among objects in the Morphbank system and external objects, and the ways that these connections illustrate important research concepts. |

download presentation: ppt |
|
Poster created by Katja Seltmann et al. for the 2007 tdwg meeting.
Title: Encouraging Users to Share Biodiversity Information Authors: Katja Seltmann, Greg Riccardi, Austin Mast, Fredrik Ronquist, Neelima Jammingumpula, Karolina Maneva-Jakimoska, Steve Winner and Deborah Paul (Florida State University), and Andrew Deans (North Carolina State University). Abstract: The Morphbank research project (http://morphbank.net) has created a repository for images that are useful to biodiversity researchers. The project adds significant value to stored images by managing complex metadata, by organizing images for searching, and by allowing users\' comments and annotations to be directly linked to images and metadata. Its sophisticated user interfaces allow users to identify new taxa, characters and matrices, and ad hoc collections of images and other objects. The productive and dedicated R&D team has helped researchers organize and use their images productively on the Web. Morphbank has more than one terabyte of images covering a broad spectrum of life. It has users and images from diverse collections, taxonomic groups and research projects. This broad user base has stressed the need for flexibility in the Morphbank system. To succeed in attracting users, recognition of the effort it takes for scientists to participate in Web dissemination activities must be acknowledged in biology research circles. Some examples of helpful strategies are: Credit for service. Each user logo, a list of contributors, and links to major projects are prominently displayed. All are useful in helping to promote the biologists work and to provide evidence of scholarly activity. Big results with little effort. Many of the new users to the system are attracted by the ability to link collections to publications. It is these big results with little effort that will persuade users to participate. Helping users learn. Straightforward user manuals, workshops, and a contact phone number help users learn the system. Many need someone to consult and it is important for users to feel welcome. Workshops may also include discussions of the value of data beyond the scope of the researchers\' individual projects, and provide important feedback for the Morphbank R&D team. Need for control. Biologists who are not skilled in programming often feel controlled by programmers or left out of the process. The differences in the way programmers and biologists approach issues may lead to difficulties. For example, biologists wish to freely edit data in the system, but developers worry that data structure and the ability to modify and augment data needs to be protected and controlled. Biologists agree with this but believe they are the ones best suited to make these decisions, not the creators of the database. This poster will illustrate how Morphbank influences the user\'s of the data they are importing into the system. There is a correlation between the level of difficulty in getting the data into the system and the perceived value of the data by the biologist. This manifests itself on many levels: 1. desire to keep data hidden, 2. wanting to be sure only the best image is used (unwillingness to have imperfect data), and 3. a feeling of being overwhelmed by the technology. Generally by making access, upload and maintenance easier, users will feel they have control over the data and will be more apt to share. Thus, effective user interface design crucial to encouraging participation. |

download poster: large print pdf or jpg |
| A general poster highlighting the great image diversity found in morphbank. The poster hightlights images from a number of the projects in the system including: Drosophila morphogenetics project at Florida State University, Herbarium at the University of Alaska Museum of the North, Hymenoptera Assembling the Tree of Life , PlantCollections™, Platygastroidea Planetary Biodiversity Inventory , Robert K. Godfrey Herbarium online database project at Florida State University, Systematic Entomology Laboratory USDA-ARS, Bishop Museum tropical fish images from Ichthyologist and photographer John E. Randall, Area de Conservacion Guanacaste (ACG), Tabanid PEET project, Gail Kampmeier and the Diptera Tree of Life Project, Brian Brown at the Natural History Museum of Los Angeles County, description of Ibalia larvae by Jose Luis Nieves-Aldrey and thesis work of Martin Wiemers and Albert Prieto-Marquez A large pdf version of this poster is available for download. |
download poster: large print pdf or jpg |
|
Poster created by Albert Prieto-Marquez et al. for the 8th International Conference of Vertebrate Morphology (ICVM8). Designed to explain creation of characters and character states in morphbank. Flow chart showing the importing of an existing Nexus file to create characters in morphbank and the illustrating of a morphbank matrix with images and annotations.
The ICVM8 16-21 July 2007. Paris, France. |
download poster: pdf or jpg |
|
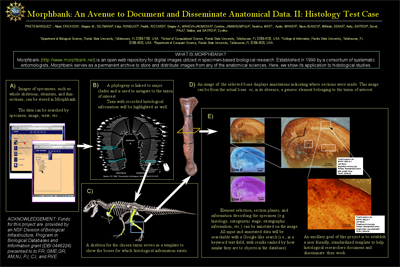
Poster created by Albert Prieto-Marquez et al. for the 8th International Conference of Vertebrate Morphology (ICVM8). This poster explains the workflow for paleohistological studies being incorporated into morphbank. These tools are under development by the morphbank team. The ICVM8 16-21 July 2007. Paris, France. |
download poster: pdf or jpg |
|
Presentation created by Katja Seltmann et al. for the 8th International Conference of Vertebrate Morphology (ICVM8). This presentation gives an introduction to morphbank tools and image management utilities. Shows screen shots of specimen, image, collection and annotation records. A good general introduction to the system.
The ICVM8 16-21 July 2007. Paris, France. |

download presentation: ppt |
|
Presentation created by Andrew R. Deans et al. for the 2006 tdwg meeting
Title: 4.5. Ontologizing Morphological Terms for Hymenoptera (Insecta) - implementing and Benefiting from a Controlled Vocabulary Authors: Andrew R Deans, Gregory A Riccardi, Fredrik Ronquist, Florida State University Abstract: Hymenoptera encompasses a large group of organisms commonly referred to as sawflies, wasps, ants and bees. This group has historically had numerous communities of researchers untangling its mysteries. Sawfly specialists, parasitoid people, ant taxonomists, bee biologists, and aculeate workers have converged on a common language that describes the morphological characteristics of these insects. Each research group has however also cultivated its own specialized terminology that may not be applicable to other hymenopterans. Complications include: (1) words that apply to structures only found in certain taxa (e.g., "cenchrus" in sawflies); (2) words that are different from terms other workers use ("parapsidal furrow" in some taxa is homologous to the "notaulus" in other taxa); (3) words that are obscure and go unused ("lunule" in microgastrine Braconidae) and (4) words that have the same spelling but are defined differently depending on the taxon (e.g., "face" in Symphyta and Aculeata is not the same as "face" in Parasitica). Ontologies serve multiple functions in a vast array of contexts - from facilitating communication between databases to standardizing the terminology used by a particular field. The hymenopterist community seeks to ontologize the vast array of morphological terminology by: (1) carefully reviewing the literature pertaining Hymenoptera morphology; (2) systematically designating synonyms that are either valid or obsolete; (3) defining relationships between terms and (4) illustrating and verbally defining valid terms. Ontology development and editing: Several software packages are available for developing and editing ontologies, e.g. OBO-edit (Gene Ontology Consortium) and Protege-OWL (Stanford), which can export the ontology in several formats. The Hymenoptera community will likely employ a user-friendly, customizable database built using Ruby on Rails/MySQL (mx; http://hymenoptera.tamu.edu/mx/). This strategy allows more Hymenoptera morphologists to participate since there is no new software to learn, and it allows the ontology to be built remotely, from anywhere with internet access. After the ontology matures, the database will be assembled by our bioinformatics colleagues into a usable ontology. The standards and hierarchical nature of an ontology will prove invaluable when applied to resources such as MorphBank (http://www.morphbank.net/): Situation 1 - When searching for "mesosoma" images, one could potentially miss images that were tagged as "scutellum". By incorporating the ontology into the search algorithm one could return images of "mesosoma" and all related terms (e.g., terms that are "part of" the mesosoma, such as the scutellum). Situation 2 - Annotating uploaded images. Standard terminology is highlighted and linked and inappropriate synonyms are identified, while non-standard or misspelled terms remain unrecognized. This provides feedback about the quality of the annotation. Situation 3 - When scoring phylogenetic characters, users might define characters and states differently. An ontology will assist in homology assignments critical to phylogenetic analyses. We acknowledge support by: National Science Foundation |
download presentation: ppt |
|
Presentation created by Greg Riccardi et al. for the 2006 tdwg meeting
Title: Representing and Using Phylogenetic Characters in Morphbank Authors: Greg Riccardi, David Gaitros, and Fredrik Ronquist Florida State University Abstract: Integrating & sharing biotic information, Images The Morphbank project is a repository of biological images and related information that includes extensive user tools in Web site, at http://morphbank.net. The value of the images in the repository is increased by maintaining detailed metadata about the image and its associated specimen. The system includes bulk upload operations that make is simple to add image collections. The Morphbank development team deployed a new version of the Web site in summer 2006 that adds extensive annotation tools for image annotations and determination annotations. A determination annotation is an object in the system that associates a specimen with an assessment of the correctness of its taxon identification. A scientist can create an annotation in order to register agreement or disagreement with the current determination. In the case of disagreement, an alternative taxon is specified. Each annotation contains image annotations that identify areas of interest related to the determination. The specimens of the Florida State University Herbarium are now included in the MorphBank repository as image and specimen objects. The Morphbank determination annotation tools are being used to evaluate the determinations of the specimen. The tools will allow experts to record their assessments of the quality of the herbarium metadata and to correct the determinations as necessary. The next annotation capability that will be included in the Morphbank tools is character state annotation, the association of a phylogenetic character state with a specific area of interest of an image. Development has begun on tools to create and manipulate phylogenetic characters and their states. These tools support importing and exporting of Nexus files. Other systems, e.g. Mesquite and MorphoBank. support the creation of character matrices that associate species with character states. Morphobank provides tools to include and annotate images of the species. Character state annotations in Morphbank are focused on identifying areas within images that are show particular character states. Each image may have many annotations. Morphbank image annotations can be used to create character matrices that relate species to states, and may also be used to relate specimens to states. The search and browse capabilities of the Morphbank tools will provide scientists with capabilities to find and compare character state annotations in ways that are not currently possible. Scientists can find all images that display particular characters or states, can use image annotations to clearly identify and differentiate the states, and can easily find appropriate characters to use for their specimens. The character definitions will be shared among all users of MorphBank. Current development plans call for the character state annotation tools to be fully implemented and deployed in advance of the TDWG 06 meeting. The TDWG talk for this abstract will include illustrations of the use of both determination and character state annotations. Details of the data models that support annotations and demonstrations of the tools will also be included. |

download presentation: ppt |


